Research Highlights
Concerted research effort at the Department of Human Genetics Amsterdam UMC resulted in 327 publications in 2021. Below, two high impact scientific publications are highlighted.
A New Role for Dynamin in Neuron Signaling
SECTION: FUNCTIONAL GENOMICS

Long-lasting modulation
Neurotransmitters are chemical messages that are transmitted to a neighboring neuron for fast transmission of a signal. Neuropeptides target a group of neurons or organ cells displaying specific receptors. Neuropeptides are more indirect, especially if they involve downstream messengers, but product a long-lasting effect.
Compared to neurotransmitters, much less is known about how neuropeptides are organized and released. Humans carry a diverse array of different neuropeptides, with over 100 known and 1,000 predicted from genome analysis. They control many physiological functions such as brain development, synaptic plasticity, circadian rhythm, behavior and emotions. Defects in their signaling are associated with multiple psychiatric disorders, obesity, and diabetes.
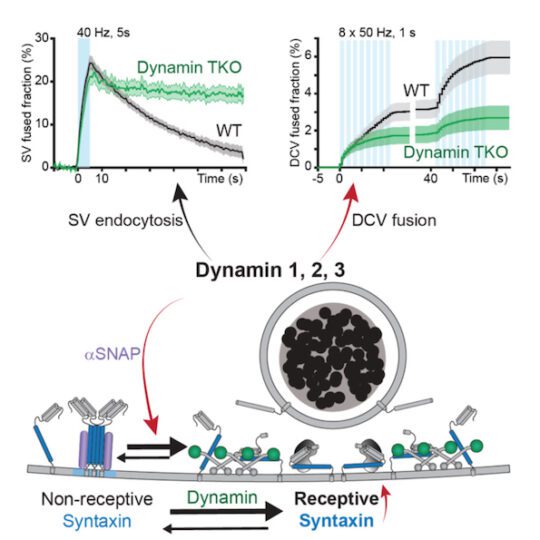
A new role for dynamin
Dr. Alessandro Moro, Anne van Nifterick, Dr. Ruud F. Toonen, and Prof. Matthijs Verhage, from the Center for Neurogenomics and Cognitive Research in the Department of Human Genetics, reported the discovery that the protein dynamin is a central regulator for neuromodulator secretion from dense core vesicles in mammalian neurons. The study, published in Science Advances in May 2021, reveals a surprising new role for dynamin, which was known to be involved in bringing dense core vesicles into the cell (endocytic), but not for also sending them out (exocytosis).
Dr. Alessandro Moro explains: “Humans have three types of dynamins. Genetic or pharmacological inactivation of all three dynamins strongly impaired dense core vesicle exocytosis but did not affect the other synaptic vesicle pathway. Wildtype dynamin restored normal exocytosis, but not GTPase-deficient or membrane-binding mutants that cause neurodevelopmental syndromes.”
The authors propose the newly discovered role of dynamin is unique from its well-characterized role in endocytosis. “Dynamins appear to have two crucial roles in dense core vesicle exocytosis: They organize the vesicle fusion sites and, in addition, recover or reactivate fusion sites for repeated use.”
These insights contribute to the current knowledge of the biological mechanisms underlying some neurodevelopmental syndromes.
Discovery of New Genes in DNA Damage Repair with CRISPR
SECTION: ONCOGENETICS
Researchers Klaas de Lint and Dr. Rob Wolthuis introduced a full-genome CRISPR functional genetic screening pipeline in Cas9-inducible diploid cells to identify genes involved in DNA damage responses. In collaboration with the Dr. Martijn Luijsterburg lab (LUMC) they first focused on identifying the human genes involved in the process of Transcription Coupled DNA Repair. Their discoveries were published in Nature Cell Biology in June 2021.

With the Nobel prize-winning application of CRISPR technology in human cells, it recently became possible for researchers to systematically disrupt human genes in lab-grown cells to reveal their roles in cancer relevant biological processes.
ELOF1 is particularly important to help cells deal with UV and chemotherapeutic compounds, but also plays a role in controlled cell division. These findings increase our understanding of hereditary cancer syndromes which are linked to DNA damage repair genes, as well as the effects of targeted drugs used in cancer therapy.
The Wolthuis lab implemented a new pipeline for so-called pooled lentiviral full-genome CRISPR-Cas9 knockout screening. A collection of 71,000 different CRISPR-lentiviruses was used to create a population of millions of cells, each containing a knockout of a unique human gene. Subsequently it was measured which genes are essential for cell growth and survival, before or after treatment with a specific DNA-damaging drug. With this technique, the researchers were able to pinpoint the human genes and pathways involved in the underlying DNA repair pathway of this study, called Transcription-Coupled DNA Repair (TCR).
Apart from all the known TCR genes, the previously uncharacterized ELOF1 gene was identified as a top hit. Next, a wide range of experimental approaches was used, including follow-up drug screens in isogenic cell lines to elucidate the precise function of ELOF1. The researchers discovered that ELOF1 is a novel component of the mRNA synthesis machinery (called the RNA-polymerase II complex), but also acts as an important factor in DNA repair when the RNA-polymerase II complex encounters a transcription-blocking lesion.
Read the publication here: Van der Weegen, Y., De Lint, K., Van den Heuvel, D. et al. (2021) ELOF1 is a transcription-coupled DNA repair factor that directs RNA polymerase II ubiquitylation. Nat. Cell Biol. 23, 595–60.https://doi.org/10.1038/s41556-021-00688-9